Bayesian inference with Turing.jl
Compatibility with the Distributions.jl API allows extensive interaction with the broader Julia ecosystem. One of the first examples discovered was the possibility to perform Bayesian inference of model parameters (including copulas) with Turing.jl.
Consider that we have a given model with a certain copula and certain marginals, all having parameters to be fitted. Then we can use Turing's @addlogprob! to compute the loglikelihood of our model and maximize it around the parameters alongside the chain as follows:
using Copulas
using Distributions
using Random
using Turing
using StatsPlots
Random.seed!(123)
true_θ = 7
true_θ₁ = 1
true_θ₂ = 3
true_θ₃ = 2
D = SklarDist(ClaytonCopula(3,true_θ), (Exponential(true_θ₁), Pareto(true_θ₂), Exponential(true_θ₃)))
draws = rand(D, 2000)
@model function copula(X)
# Priors
θ ~ TruncatedNormal(1.0, 1.0, -1/3, Inf)
θ₁ ~ TruncatedNormal(1.0, 1.0, 0, Inf)
θ₂ ~ TruncatedNormal(1.0, 1.0, 0, Inf)
θ₃ ~ TruncatedNormal(1.0, 1.0, 0, Inf)
# Build the parametric model
C = ClaytonCopula(3,θ)
X₁ = Exponential(θ₁)
X₂ = Pareto(θ₂)
X₃ = Exponential(θ₃)
D = SklarDist(C, (X₁, X₂, X₃))
# Compute the final loglikelyhood
Turing.Turing.@addlogprob! loglikelihood(D, X)
end
sampler = NUTS() # MH() works too
chain = sample(copula(draws), sampler, MCMCThreads(), 100, 4)Note that we truncated the θ parameter at -1/3 and not 0, as the ClaytonCopula can handle negative dependence structures. Only 100 steps were run for efficiency; you can increase this number as needed. The code above outputs a summary of the chain :
Summary Statistics:
| parameters | true value | mean | std | mcse | ess_bulk | ess_tail | rhat | ess_per_sec |
|---|---|---|---|---|---|---|---|---|
| Symbol | Float64 | Float64 | Float64 | Float64 | Float64 | Float64 | Float64 | Float64 |
| θ | 7.0 | 6.9319 | 0.1150 | 0.0067 | 291.8858 | 267.1353 | 1.0061 | 0.7238 |
| θ₁ | 1.0 | 0.9954 | 0.0209 | 0.0019 | 116.6941 | 94.3070 | 1.0347 | 0.2894 |
| θ₂ | 3.0 | 2.9839 | 0.0639 | 0.0062 | 108.9185 | 105.5284 | 1.0390 | 0.2701 |
| θ₃ | 2.0 | 2.0055 | 0.0418 | 0.0039 | 114.7324 | 109.5396 | 1.0328 | 0.2845 |
Quantiles:
| parameters | 2.5% | 25.0% | 50.0% | 75.0% | 97.5% |
|---|---|---|---|---|---|
| Symbol | Float64 | Float64 | Float64 | Float64 | Float64 |
| θ | 6.7286 | 6.8438 | 6.9330 | 7.0150 | 7.1436 |
| θ₁ | 0.9555 | 0.9818 | 0.9953 | 1.0093 | 1.0386 |
| θ₂ | 2.8606 | 2.9426 | 2.9859 | 3.0196 | 3.1186 |
| θ₃ | 1.9254 | 1.9758 | 2.0056 | 2.0336 | 2.0923 |
And then plot(chain) produces the following plot:
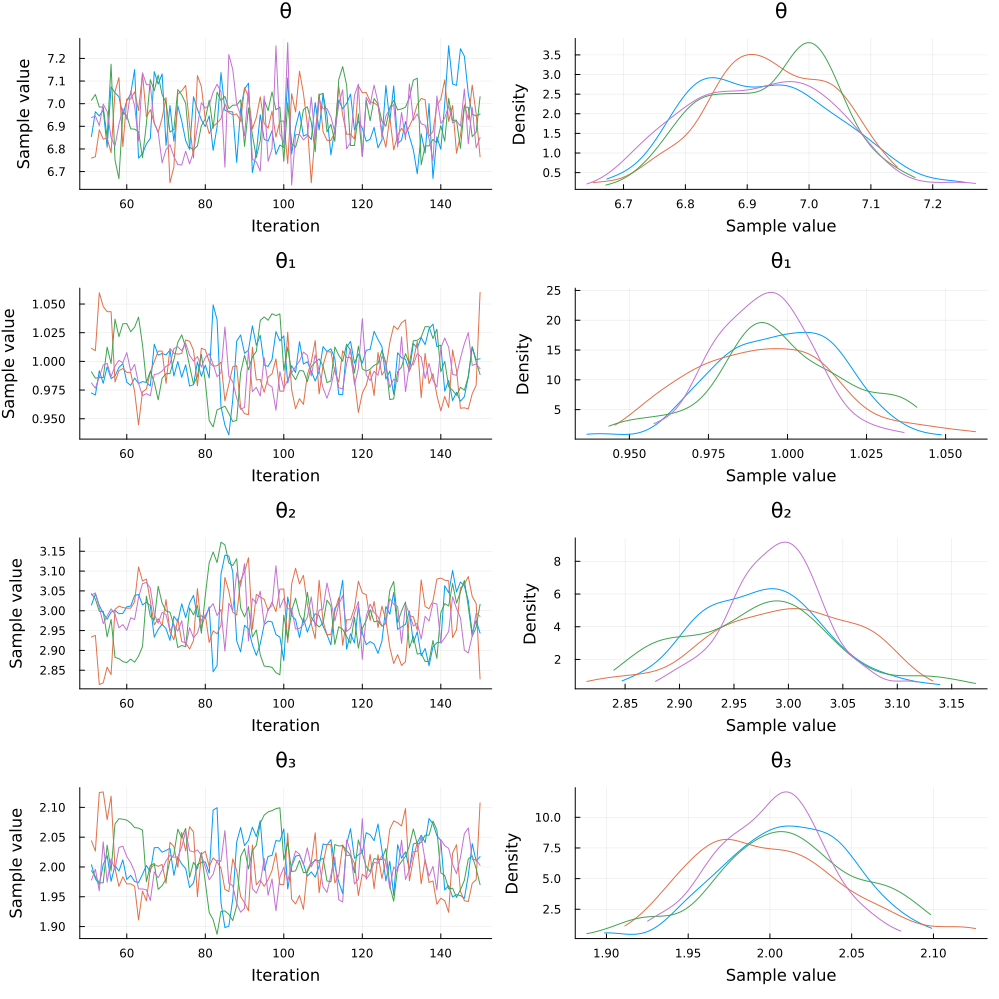
Similar approaches can be used to fit many other dependence structures in a Bayesian setting. The example above showcases parametric estimation of a sampling model, but Bayesian regression with an error structure given by a parametric copula is just as easy to implement.
This was run on the following environment:
julia> versioninfo()
Julia Version 1.10.0
Commit 3120989f39 (2023-12-25 18:01 UTC)
[...]
(env) pkg> status
[ae264745] Copulas v0.1.20
[31c24e10] Distributions v0.25.107
[f3b207a7] StatsPlots v0.15.6
[fce5fe82] Turing v0.30.3
[9a3f8284] Random