Conditioning and Subsetting
Conditioning
This page introduces conditional distributions under a copula model and shows how to construct them programmatically using condition. The same interface works either on the uniform scale (copula only) or on the original scale (via SklarDist).
Overview
Take a D-variate copula
which is a proper distribution on
where u at index i and 1 elsewhere in I.
On the original scale for a compound distribution X = SklarDist(C, (X_1,…,X_D)), conditioning on values
The copula of the conditional vector ConditionalCopula(C, J, u_J) and used internally by condition. The condition function can be used as follows:
condition(C::Copula, js, u_js)returns the conditional distribution on the uniform scale forI = setdiff(1:D, js). Iflength(I) == 1, the result is a univariate distribution supported on, subclass of Distortion, and otherwise it is aSklarDist(::ConditionalCopula, NTuple{d,<:Distortion}).condition(X::SklarDist, js, x_js)returns the conditional distribution on the original scale by pushing forward each distortion through the corresponding marginal.For known parametric families, there are fast paths implemented mostly as subclass to
DistortionsorConditionalCopula, but this should be completely transparent to the user.
If you find a conditional that should admit a faster closed-form or semi-analytic path but currently falls back to the generic construction, please open an issue, we’ll happily implement it 😃
Examples
Let us visualize a given univariate distortion:
using Copulas, Distributions, Plots, StatsBase
C = ClaytonCopula(2, 1.5)
D = condition(C, 2, 0.3) # distortion for U₁ | U₂ = 0.3
ts = range(0.0, 1.0; length=401)
plt = plot(ts, cdf.(Ref(D), ts);
xlabel="u", ylabel="H_{1|2}(u | 0.3)",
title="Conditional CDF on the uniform scale",
legend=false)
plt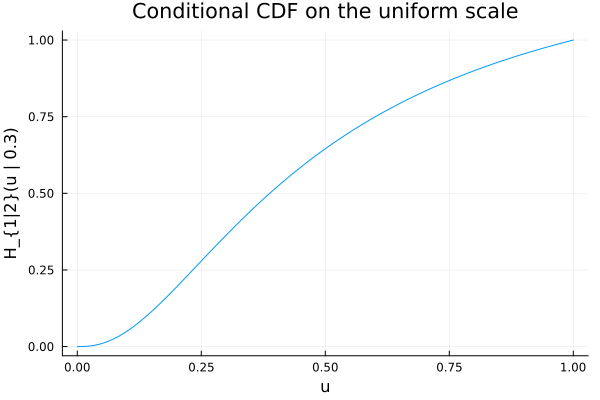
Confirm the result by overlaying the empirical cdf of a sample:
N = 2000
αs = rand(N)
us = Distributions.quantile.(Ref(D), αs)
ECDF = ecdf(us)
plot!(ts, ECDF.(ts); seriestype=:steppost, label="empirical", alpha=0.6, color=:black)
plot!(ts, cdf.(Ref(D), ts); label="analytic", color=:blue)
plt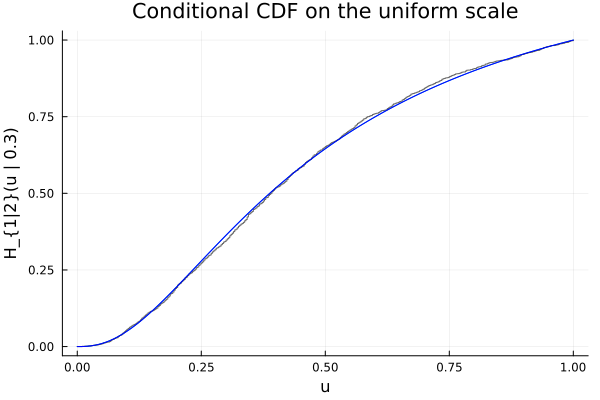
The same thing can be done on marginal scales using SklarDist:
C = ClaytonCopula(2, 1.5)
X = SklarDist(C, (Normal(), Normal()))
X1_given_X2 = condition(X, 2, 0.0) # distribution of X₁ | X₂ = 0.0
cdf(X1_given_X2, 1.0), quantile(X1_given_X2, 0.95)(0.8472337930319087, 1.5990201973152853)xs = rand(X1_given_X2, 2000)
Fx = ecdf(xs)
xs_grid = range(quantile(X1_given_X2, 0.001), quantile(X1_given_X2, 0.999); length=401)
plot(xs_grid, Distributions.cdf.(Ref(X1_given_X2), xs_grid);
xlabel="x", ylabel="F_{X₁|X₂}(x|0)", title="Original-scale conditional CDF", label="analytic")
plot!(xs_grid, Fx.(xs_grid); seriestype=:steppost, label="empirical", alpha=0.6, color=:black)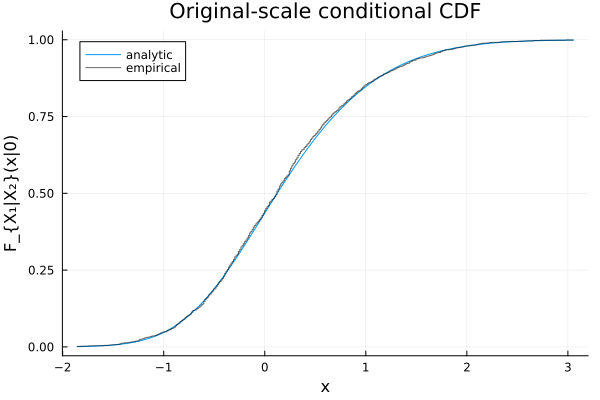
When conditioning on less than SklarDist:
H = condition(ClaytonCopula(4, 4.2), (2, 3), (0.25, 0.8))SklarDist{ArchimedeanCopula{2, TiltedGenerator{Copulas.ClaytonGenerator{Float64}, Float64, 2}}, Tuple{Copulas.ArchimedeanDistortion{Copulas.ClaytonGenerator{Float64}, Float64, 2}, Copulas.ArchimedeanDistortion{Copulas.ClaytonGenerator{Float64}, Float64, 2}}}(
C: ArchimedeanCopula{2, TiltedGenerator{Copulas.ClaytonGenerator{Float64}, Float64, 2}}(θ = 4.2, sJ = 80.55877526114394)
m: (Copulas.ArchimedeanDistortion{Copulas.ClaytonGenerator{Float64}, Float64, 2}(
G: Copulas.ClaytonGenerator{Float64}(4.2)
sJ: 80.55877526114394
den: 1.1276684721576129e-5
)
, Copulas.ArchimedeanDistortion{Copulas.ClaytonGenerator{Float64}, Float64, 2}(
G: Copulas.ClaytonGenerator{Float64}(4.2)
sJ: 80.55877526114394
den: 1.1276684721576129e-5
)
)
)plot(H)
Relation to the conditional copula
The conditional copula ConditionalCopula(C, js, u_js) and is used as the copula of the conditional joint when |I| > 1. When condition returns a SklarDist (i.e., when |I| > 1), you can access this copula directly via the .C field of the returned object:
H.C # the copulaArchimedeanCopula{2, TiltedGenerator{Copulas.ClaytonGenerator{Float64}, Float64, 2}}(θ = 4.2, sJ = 80.55877526114394)H.m # the marginals(Copulas.ArchimedeanDistortion{Copulas.ClaytonGenerator{Float64}, Float64, 2}(
G: Copulas.ClaytonGenerator{Float64}(4.2)
sJ: 80.55877526114394
den: 1.1276684721576129e-5
)
, Copulas.ArchimedeanDistortion{Copulas.ClaytonGenerator{Float64}, Float64, 2}(
G: Copulas.ClaytonGenerator{Float64}(4.2)
sJ: 80.55877526114394
den: 1.1276684721576129e-5
)
)Implementation
Copulas.condition Function
condition(C::Copula{D}, js, u_js)
condition(X::SklarDist, js, x_js)Construct conditional distributions with respect to a copula, either on the uniform scale (when passing a Copula) or on the original data scale (when passing a SklarDist).
Arguments
C::Copula{D}: D-variate copulaX::SklarDist: joint distribution with copulaX.Cand marginalsX.mjs: indices of conditioned coordinates (tuple, NTuple, or vector)u_js: values in [0,1] forU_js(when conditioning a copula)x_js: values on original scale forX_js(when conditioning a SklarDist)j, u_j, x_j: 1D convenience overloads for the common p = 1 case
Returns
If the number of remaining coordinates
d = D - length(js)is 1:condition(C, js, u_js)returns aDistortionon [0,1] describingU_i | U_js = u_js.condition(X, js, x_js)returns an unconditional univariate distribution forX_i | X_js = x_js, computed as the push-forwardD(X.m[i])whereD = condition(C, js, u_js)andu_js = cdf.(X.m[js], x_js).
If
d > 1:condition(C, js, u_js)returns the conditional joint distribution on the uniform scale as aSklarDist(ConditionalCopula, distortions).condition(X, js, x_js)returns the conditional joint distribution on the original scale as aSklarDistwith copulaConditionalCopula(C, js, u_js)and appropriately distorted marginalsD_k(X.m[i_k]).
Notes
For best performance, pass
jsandu_jsas NTuple to keepp = length(js)known at compile time. The specialized methodcondition(::Copula{2}, j, u_j)exploits this for the commonD = 2, d = 1case.Specializations are provided for many copula families (Independent, Gaussian, t, Archimedean, several bivariate families). Others fall back to an automatic differentiation based construction.
This function returns the conditional joint distribution
H_{I|J}(· | u_J). The “conditional copula” isConditionalCopula(C, js, u_js), i.e., the copula of that conditional distribution.
Copulas.Distortion Type
Distortion <: Distributions.ContinuousUnivariateDistributionAbstract super-type for objects describing the (uniform-scale) conditional marginal transformation U_i | U_J = u_J of a copula.
Subtypes implement cdf/quantile on [0,1]. They are not full arbitrary distributions; they model how a uniform variable is distorted by conditioning. They can be applied as a function to a base marginal distribution to obtain the conditional marginal on the original scale: if D::Distortion and X::UnivariateDistribution, then D(X) is the distribution of X_i | U_J = u_J.
Copulas.DistortionFromCop Type
DistortionFromCop{TC,p,T} <: DistortionGeneric, uniform-scale conditional marginal transformation for a copula.
This is the default fallback (based on mixed partial derivatives computed via automatic differentiation) used when a faster specialized Distortion is not available for a given copula family.
Parameters
TC: copula typep: length of the conditioned index set J (static)T: element type for the conditioned values u_J
Construction
DistortionFromCop(C::Copula, js::NTuple{p,Int}, ujs::NTuple{p,<:Real}, i::Int)builds the distortion for the conditional marginal of indexigivenU_js = ujs.
Notes
- A convenience method
DistortionFromCop(C, j::Int, uj::Real, i::Int)exists for the commonp = 1case.
Copulas.DistortedDist Type
DistortedDist{Disto,Distrib} <: Distributions.UnivariateDistributionPush-forward of a base marginal by a Distortion.
Copulas.ConditionalCopula Type
ConditionalCopula{d} <: Copula{d}Copula of the conditioned random vector U_I | U_J = u_J.
sourceSee also
condition— reference documentation with all calling syntaxesSklarDist— compound distributions via Sklar’s theoremrosenblatt— sequential transforms (related but different)
Subsetting
Subsetting extracts the dependence structure among a subset of coordinates. Given a copula C of dimension d and an index tuple dims::NTuple{p,Int}, the function subsetdims returns a copula on those p dimensions that preserves the original dependence restricted to dims.
There are two entry points:
subsetdims(C::Copula, dims)returns aCopula{p}(orUniform()whenp == 1).subsetdims(X::SklarDist, dims)returns aSklarDistwith copulasubsetdims(C, dims)and marginals(m[i] for i in dims).
Internally, we materialize subsetting with a small wrapper type SubsetCopula{p}(C, dims) which delegates cdf, pdf, and sampling to the base copula by saturating non-selected coordinates at 1. For many families we provide specialized constructors that return the natural reduced-parameter form instead of a wrapper (e.g., elliptical copulas return the appropriate submatrix, Archimedean keeps the same generator with reduced dimension, etc.). It can be used as follows:
using Copulas, Distributions
C = GaussianCopula([1.0 0.6 0.2; 0.6 1.0 0.3; 0.2 0.3 1.0])
S = subsetdims(C, (1,3)) # 2D copula on coordinates 1 and 3
length(S), typeof(S)(2, GaussianCopula{2, Matrix{Float64}})X = SklarDist(C, (Normal(), Normal(1,2), LogNormal()))
X13 = subsetdims(X, (1,3)) # keeps marginals (Normal(), LogNormal()) and reduces the copula
length(X13.C), length(X13.m)(2, 2)The resulting object depends on the copula familly, since some fast paths are given. If no specialization exists, a SubsetCopula wrapper is returned. It’s fully usable and equivalent from an API perspective; specialized forms simply yield better performance and clearer display.
Subsetting and conditioning commute in the obvious way: conditioning on coordinates J and then extracting a subset of the remaining coordinates is equivalent to subsetting the base copula first and then conditioning on the corresponding indices. In code, if S = subsetdims(C, dims), conditioning on indices js within S is implemented by mapping js to indices in the base copula and delegating to ConditionalCopula(C, ·, ·); the resulting conditional copula of S is either the base conditional copula (when all remaining coordinates are kept) or a further SubsetCopula of it.
Examples
# Archimedean example
C = ClaytonCopula(3, 2.0)
S = subsetdims(C, (1,2)) # still a ClaytonCopula with the same parameter
rand(S, 3) # sample 3 points
cdf(S, [0.7, 0.9])0.6629375642933931# Survival example with flips remapped
base = GaussianCopula([1.0 0.7 0.2; 0.7 1.0 0.1; 0.2 0.1 1.0])
S = SurvivalCopula(base, (2,))
S13 = subsetdims(S, (1,3)) # flip on 2 drops; no flips remain
typeof(S13), S13 isa SurvivalCopula(GaussianCopula{2, Matrix{Float64}}, false)Implementation
Copulas.subsetdims Function
subsetdims(C::Copula, dims::NTuple{p, Int})
subsetdims(D::SklarDist, dims)Return a new copula or Sklar distribution corresponding to the subset of dimensions specified by dims.
Arguments
C::Copula: The original copula object.D::SklarDist: The original Sklar distribution.dims::NTuple{p, Int}: Tuple of indices representing the dimensions to keep.
Returns
- A
SubsetCopulaor a newSklarDistobject corresponding to the selected dimensions. Ifp == 1, returns aUniformdistribution or the corresponding marginal.
Details
This function extracts the dependence structure among the specified dimensions from the original copula or Sklar distribution. Specialized methods exist for some copula types to ensure efficiency and correctness.
sourceCopulas.SubsetCopula Type
SubsetCopula{d,CT}Fields:
C::CT- The copuladims::Tuple{Int}- a Tuple representing which dimensions are used.
Constructor
SubsetCopula(C::Copula,dims)This class allows to construct a random vector corresponding to a few dimensions of the starting copula. If C, this returns the copula of ( for i in dims). The dependence structure is preserved. There are specialized methods for some copulas.
Rosenblatt transformations
Definition and usefulness
The Rosenblatt transformation considers a random vector
More formally, consider the map
References:
[13] Rosenblatt, M. (1952). Remarks on a multivariate transformation. Annals of Mathematical Statistics, 23(3), 470-472.
[4] Joe, H. (2014). Dependence Modeling with Copulas. CRC Press. (Section 2.10)
[14] McNeil, A. J., & Nešlehová, J. (2009). Multivariate Archimedean copulas, d-monotone functions and ℓ 1-norm symmetric distributions.
In certain circumstances, in particular for Archimedean copulas, this map simplifies to tractable expressions. It has a few nice properties:
is a bijection.
These two properties are leveraged in some cases to construct the inverse Rosenblatt transformations, which map random noise to proper samples from the copula. In some cases, this is the best sampling algorithm available.
Implementation
As soon as the random vector X that subtypes SklarDist or Copula, you have access to the rosenblatt(X, x) and inverse_rosenblatt(X, x) operators, which both have a straightforward interpretation from their names.
Copulas.rosenblatt Function
rosenblatt(C::Copula, u)Computes the rosenblatt transform associated to the copula C on the vector u. Formally, assuming that U ∼ C, the result should be uniformely distributed on the unit hypercube. The importance of this transofrmation comes from its bijectivity: inverse_rosenblatt(C, rand(d)) is equivalent to rand(C). The interface proposes faster versions for matrix inputs u.
Generic Rosenblatt transform using conditional distortions: S₁ = U₁, S_k = H_{k|1:(k-1)}(U_k | U₁:U_{k-1}). Specialized families may provide faster overrides.
[13] Rosenblatt, M. (1952). Remarks on a multivariate transformation. Annals of Mathematical Statistics, 23(3), 470-472.
[4] Joe, H. (2014). Dependence Modeling with Copulas. CRC Press. (Section 2.10)
[14] McNeil, A. J., & Nešlehová, J. (2009). Multivariate Archimedean copulas, d-monotone functions and ℓ 1-norm symmetric distributions.
Copulas.inverse_rosenblatt Function
inverse_rosenblatt(C::Copula, u)Computes the inverse rosenblatt transform associated to the copula C on the vector u. Formally, assuming that U ∼ Π, the independence copula, the result should be distributed as C. Also look at rosenblatt(C, u) for the inverse transformation. The interface proposes faster versions for matrix inputs u.
Generic inverse Rosenblatt using conditional distortions: U₁ = S₁, U_k = H_{k|1:(k-1)}^{-1}(S_k | U₁:U_{k-1}). Specialized families may provide faster overrides.
References:
[13] Rosenblatt, M. (1952). Remarks on a multivariate transformation. Annals of Mathematical Statistics, 23(3), 470-472.
[4] Joe, H. (2014). Dependence Modeling with Copulas. CRC Press. (Section 2.10)
[14] McNeil, A. J., & Nešlehová, J. (2009). Multivariate Archimedean copulas, d-monotone functions and ℓ 1-norm symmetric distributions.
Once again, since the rosenblatt transform leverages the conditioning mechanisme, some fast-paths might be missing in the implementation.
Sanity check plot
You can validate that the Rosenblatt transform maps samples to independent uniforms by checking the marginal ECDFs against the 45° line.
using Copulas, Plots, StatsBase
# pick a nontrivial copula
C = ClaytonCopula(3, 1.5)
# draw samples and apply Rosenblatt transform coordinate-wise
U = rand(C, 3000) # size (3, N)
S = reduce(hcat, (rosenblatt(C, U[:, i]) for i in 1:size(U, 2))) # size (3, N)
ts = range(0.0, 1.0; length=401)
layout = @layout [a b c]
plt = plot(layout=layout, size=(900, 280), legend=false)
for k in 1:3
Ek = ecdf(S[k, :])
plot!(plt[k], ts, Ek.(ts); seriestype=:steppost, color=:black,
title="ECDF of $(k)", xlabel="u", ylabel="ECDF")
plot!(plt[k], ts, ts; color=:blue, alpha=0.7)
end
plt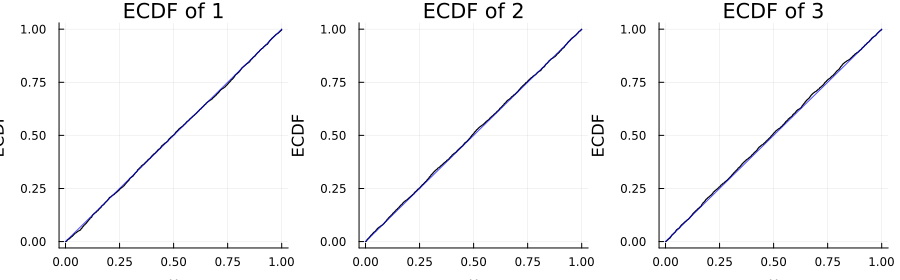
References
H. Joe. Dependence Modeling with Copulas (CRC press, 2014).
M. Rosenblatt. Remarks on a multivariate transformation. Annals of Mathematical Statistics 23, 470–472 (1952).
A. J. McNeil and J. Nešlehová. Multivariate Archimedean Copulas, d -Monotone Functions and L1 -Norm Symmetric Distributions. The Annals of Statistics 37, 3059–3097 (2009).