Nonparametric estimation of the radial law in Archimedean copulas
Introduction
This example recalls and implements a practical, nonparametric way to estimate the radial (mixing) distribution R that underlies a d-Archimedean copula by inverting a link between the kendall distribution and the radial part [41] [42], and applying this inversion to the empirical Kendall distribution. Suprisingly, a triangular recursion appears that maps empirical kendall atoms to radial atoms: see, in particular, [36]. We’ll:
explain the idea,
implement a small estimator,
validate it visually on discrete and continuous radial laws,
and compare original vs fitted copulas.
For a d-Archimedean copula with generator
The empirical Kendall distribution of the copula sample carries enough information to reconstruct a discrete approximation of
Note that there exists other approaches at archimedean estimation:
Parametric estimation fixes a generator family (Clayton, Gumbel, Frank, Joe, …) and estimates parameters by (pseudo-)likelihood, IFM, or moments such as inversion of Kendall’s
[75]; [76]. Efficient but not nonparametric. Semiparametric approaches keep a parametric generator with nonparametric margins [77].
Kendall-process methods, notably [78] for
, highlight the central role of the Kendall distribution. Extensions include settings with censoring [79].
Theoretical setup
Let
If
For empirical work, given a sample
When
Finally, it is known from [41] that when C is archimedean, the true kendall function writes
and therefore
Proposed estimator (triangular inversion)
From the Kendall pseudo-sample
so that
solved by monotone 1D root finding in
The associated generator
Specificities when
When
This closed form could be used directly (even if our code does not for the moment).
Implementation
We define a few helpers to visualize and validate the fitted model, and we now use the built-in EmpiricalGenerator as the estimator:
simulate Archimedean copula samples from a given R,
quick diagnostic plots, and
optional visualization of φ_R for a discrete R.
using Distributions, StatsBase, Roots, QuadGK, Plots, Copulas
kendall_function(u::AbstractMatrix) = (W = Copulas._kendall_sample(u); t -> count(w -> w <= t, W) / length(W))
# Williamson d-transform φ_R for several R types
mk_ϕᵣ(R::DiscreteUnivariateDistribution, d) = let supp = support(R), w = pdf.(R, supp)
t -> sum(wi * max(1 - t/x, 0)^(d-1) for (x, wi) in zip(supp, w))
end
mk_ϕᵣ(R::ContinuousUnivariateDistribution, d) = function(t)
t == 0 && return 1 - Distributions.cdf(R, 0)
val, = quadgk(x -> pdf(R, x) * (1 - t/x)^(d-1), t, Inf; rtol=1e-6)
return val
end
mk_ϕᵣ(R::DiscreteNonParametric, d) = let supp = support(R), w = pdf.(R, support(R))
t -> sum(wi * max(1 - t/r, 0)^(d-1) for (wi, r) in zip(w, supp))
end
# Simulate d×n Archimedean copula sample from a given radial R via simplex representation
function spl_cop(R, d::Int, n::Int)
ϕᵣ = mk_ϕᵣ(R, d)
S = -log.(rand(d, n))
S ./= sum(S, dims=1)
return ϕᵣ.(S .* rand(R, n)')
end
fit_empirical_generator(u::AbstractMatrix) = EmpiricalGenerator(u)
# Visual diagnostics: Kendall overlay + original vs simulated copula + (optional) histograms of R vs R̂
function diagnose_plots(u::AbstractMatrix, Rhat; R=nothing, logged=false)
d, n = size(u)
v = spl_cop(Rhat, d, n)
K = kendall_function(u)
ϕᵣ = mk_ϕᵣ(Rhat, d)
supp = support(Rhat)
a = [ϕᵣ.(r) for r in supp]
w = pdf.(Rhat, supp)
Kᵣ(x) = sum(w .* (a .< x))
xs = range(0, 1; length=1001)
# Compute difference series once and stabilize axis if essentially flat to avoid
# PlotUtils warning: "No strict ticks found" (happens when span ~ 1e-16)
diff_vals = K.(xs) .- Kᵣ.(xs)
lo, hi = extrema(diff_vals)
span = hi - lo
# Threshold below which we consider the curve numerically flat
if span < 1e-9
# Center around mean (≈ mid of lo/hi) and impose a symmetric small band
mid = (lo + hi) / 2
pad = max(span, 1e-9) # ensure non‑zero
lo_plot = mid - pad
hi_plot = mid + pad
# Provide explicit ticks so PlotUtils doesn't search for strict ticks
tickvals = (mid - pad, mid, mid + pad)
p1 = plot(xs, diff_vals; title="K̂ₙ(x) - K_R̂(x)", xlabel="", ylabel="", legend=false,
ylims=(lo_plot, hi_plot), yticks=collect(tickvals))
else
p1 = plot(xs, diff_vals; title="K̂ₙ(x) - K_R̂(x)", xlabel="", ylabel="", legend=false)
end
p2 = plot(xs, xs .- K.(xs), label="x - K̂ₙ(x)", title="x - K̂ₙ(x) vs x - K_R̂(x)", xlabel="", ylabel="")
plot!(p2, xs, xs .- Kᵣ.(xs), label="x - K_R̂(x)")
p3 = scatter(u[1, :], u[2, :], title="Original sample (first two dims)", xlabel="", ylabel="",
alpha=0.25, msw=0, label=nothing)
p4 = scatter(v[1, :], v[2, :], title="Simulated from R̂ (first two dims)", xlabel="", ylabel="",
alpha=0.25, msw=0, label=nothing)
plots_list = [p1, p3, p2, p4]
lay, sz = (2, 2), (1100, 800)
if !isnothing(R)
r1 = rand(Rhat, 10*n)
r2 = rand(R, 10*n)
ttl1, ttl2 = "R̂ histogram", "R histogram"
if logged
r1 .= log.(r1); r2 .= log.(r2)
ttl1, ttl2 = "log(R̂) histogram", "log(R) histogram"
end
p6 = histogram(r1, title=ttl1, xlabel="", ylabel="", label=nothing, normalize=:pdf)
p5 = histogram(r2, title=ttl2, xlabel="", ylabel="", label=nothing, normalize=:pdf)
plots_list = [p1, p3, p5, p2, p4, p6]
lay, sz = (2, 3), (1100, 700)
end
plot(plots_list...; layout=lay, size=sz)
end
# Optional: visualize φ_R(t) for a discrete radial law on a grid
function plot_phiR(R::DiscreteUnivariateDistribution, d::Int; tmax=maximum(support(R)), m=400)
ϕᵣ = mk_ϕᵣ(R, d)
ts = range(0, tmax; length=m)
plot(ts, ϕᵣ.(ts), xlabel="t", ylabel="φ_R(t)", title="Williamson d-transform φ_R(t)", legend=false)
endplot_phiR (generic function with 1 method)Scale of R is not identifiable: φ_R depends on ratios t/R. We normalize the largest recovered radius to 1 without loss of generality.
Numerical illustrations
We now generate samples from several radial laws, fit R̂, and compare:
Kendall function overlays,
original vs simulated copula scatter,
optionally histograms of R vs R̂ (or their logs for heavy tails).
To keep docs fast, we use modest sample sizes (n ≈ 1000–1500). Increase locally if you want smoother curves.
1) Dirac at 1 (lower-bound Clayton), d = 3
R = Dirac(1.0)
d, n = 3, 1000
u = spl_cop(R, d, n)
Ghat = EmpiricalGenerator(u)
Rhat = Copulas.𝒲₋₁(Ghat, d)
diagnose_plots(u, Rhat; R=R)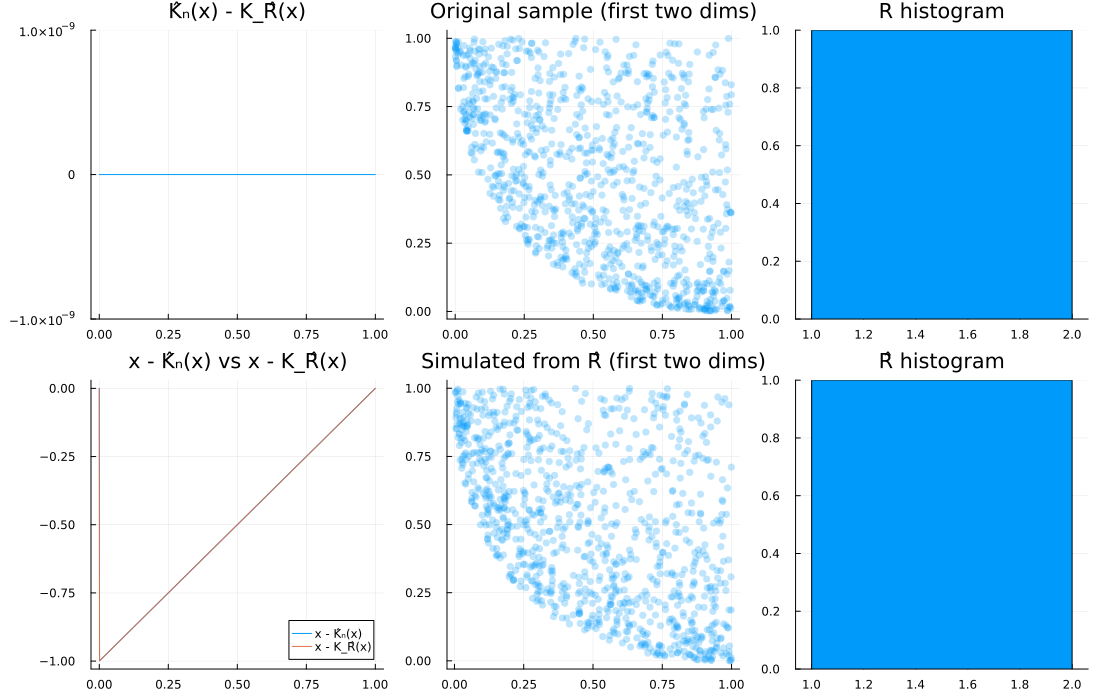
2) Mixture of point masses, d = 2
R = DiscreteNonParametric([1.0, 4.0, 8.0], fill(1/3, 3))
d, n = 2, 1000
u = spl_cop(R, d, n)
Ghat = EmpiricalGenerator(u)
Rhat = Copulas.𝒲₋₁(Ghat, d)
diagnose_plots(u, Rhat; R=R)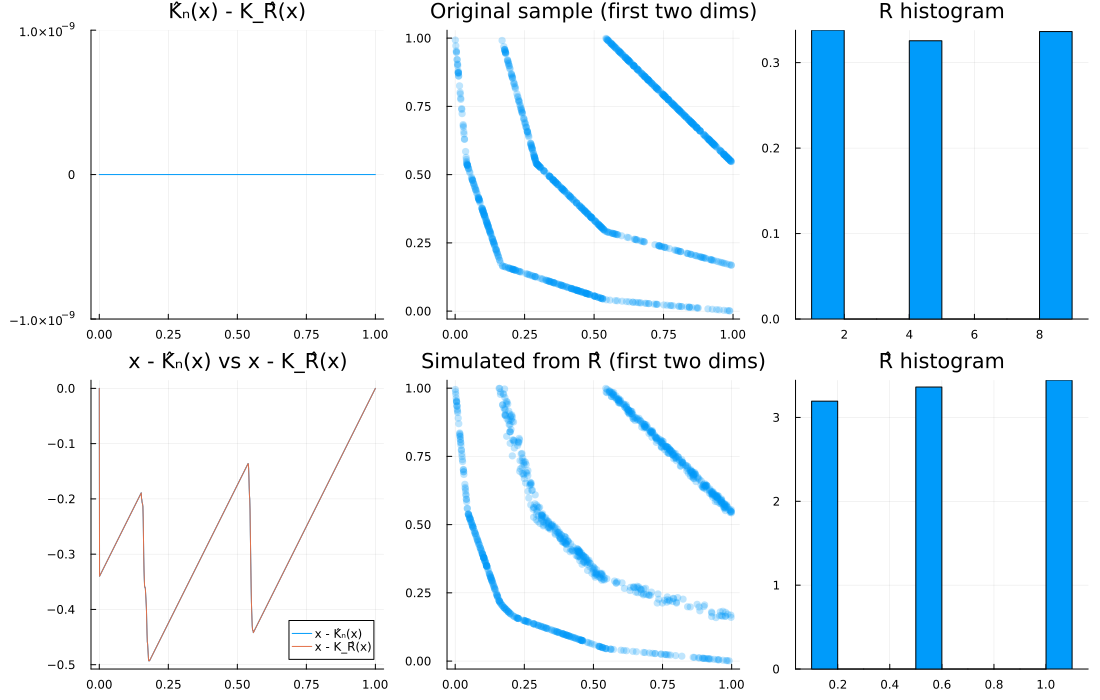
3) Mixture of point masses, d = 3
R = DiscreteNonParametric([1.0, 4.0, 8.0], fill(1/3, 3))
d, n = 3, 1000
u = spl_cop(R, d, n)
Ghat = EmpiricalGenerator(u)
Rhat = Copulas.𝒲₋₁(Ghat, d)
diagnose_plots(u, Rhat; R=R)
4) LogNormal(1, 3) (heavy tail), d = 10
R = LogNormal(1, 3)
d, n = 10, 1000
u = spl_cop(R, d, n)
Ghat = EmpiricalGenerator(u)
Rhat = Copulas.𝒲₋₁(Ghat, d)
diagnose_plots(u, Rhat; R=R, logged=true)
5) Pareto(1/2) (infinite mean), d = 10
R = Pareto(1.0, 1/2)
d, n = 10, 1000
u = spl_cop(R, d, n)
Ghat = EmpiricalGenerator(u)
Rhat = Copulas.𝒲₋₁(Ghat, d)
diagnose_plots(u, Rhat; R=R, logged=true)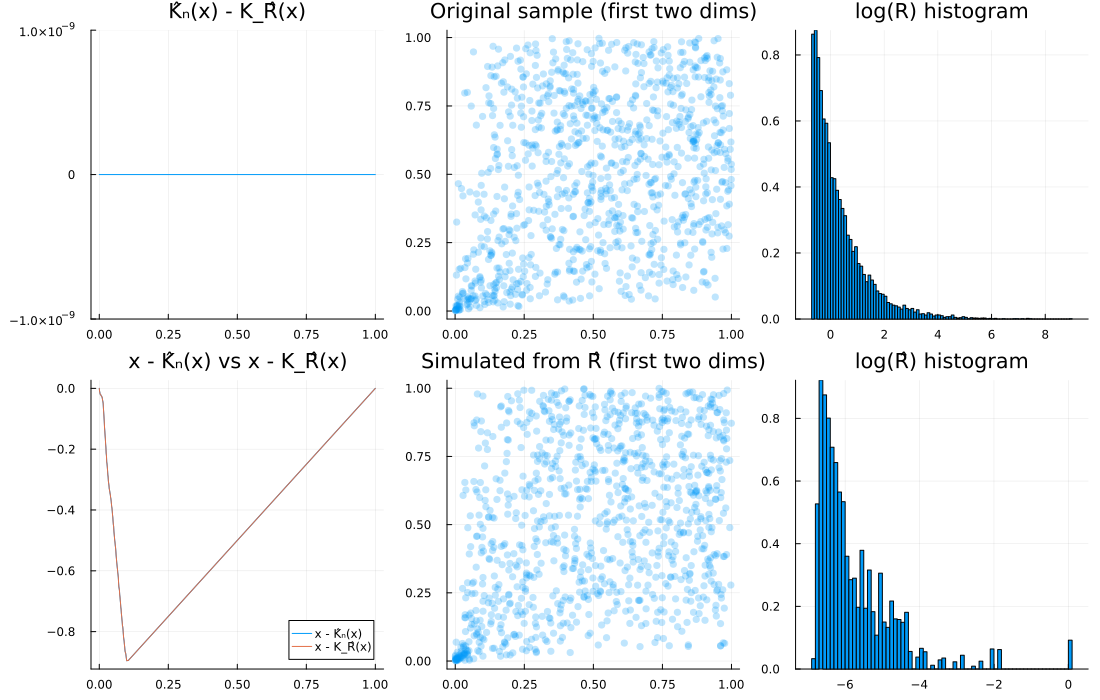
A quick look at
We can visualize
Rex = DiscreteNonParametric([0.2, 0.5, 1.0], [0.2, 0.3, 0.5])
plot_phiR(Rex, 3)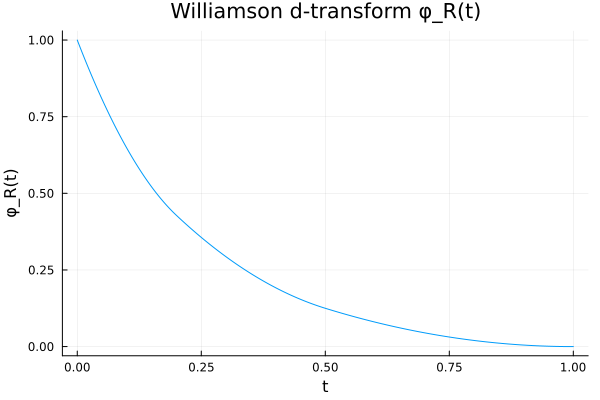
Discussion and caveats
The fit is discrete even when the true
is continuous: that’s expected. As grows, atoms densify in the bulk where Kendall jumps concentrate. Scale is not identifiable; we fix
. Very small weights can make the inversion numerically sensitive. Bootstrap CIs (not shown here) can complement delta-method formulas.
Liouville extension (remark)
The decoupling between the simplex and radial part allows the same idea to estimate radial parts of Liouville copulas. If
so Kendall atoms have the same form as in the Archimedean case, and the triangular inversion applies. Estimation of the Dirichlet parameters can be handled separately.
Once Liouville copulas are implemented in the package, we will probably use this method to fit them.
Appendix: Jacobian of the recursion (might be usefull for inference).
Define, for
Partial derivatives at the solution
Differentiating
Solve downward in
The fitted generator
Combine with the chain rule to propagate variances via the delta method if needed.
References
C. Genest, J. Nešlehová and J. Ziegel. Inference in Multivariate Archimedean Copula Models. TEST 20, 223–256 (2011).
A. J. McNeil and J. Nešlehová. Multivariate Archimedean copulas,
-monotone functions and -norm symmetric distributions. Annals of Statistics 37, 3059–3097 (2009). R. E. Williamson. Multiply monotone functions and their Laplace transforms. Duke Mathematical Journal 23, 189–207 (1956).
A. J. McNeil, R. Frey and P. Embrechts. Estimation of copula models. Quantitative Risk Management: Concepts, Techniques and Tools, 235–284 (2008).
M. Hofert and A. J. McNeil. Nesting Archimedean copulas. Statistica Sinica 22, 441–477 (2012).
C. Genest, K. Ghoudi and L.-P. Rivest. A semiparametric estimation procedure of dependence parameters in multivariate families of distributions. Biometrika 82, 543–552 (1995).
C. Genest and L.-P. Rivest. Statistical inference procedures for bivariate Archimedean copulas. Journal of the American Statistical Association 88, 1034–1043 (1993).
M. Michaelides, H. Cossette and M. Pigeon. A non-parametric estimator for Archimedean copulas under flexible censoring scenarios and an application to claims reserving, arXiv preprint arXiv:2401.07724 (2024).